Introduction
The separation of proteins in the laboratory is an important step in many biochemistry techniques. One major method employed in separating these proteins is electrophoresis. SDS-PAGE electrophoresis relies on the use of a detergent known as sodium dodecyl sulfate (SDS) (Albert, David, & Michael, 2005). This detergent contains a large amount of negative charges such that when added to a protein solution, it renders all the charges in the protein null. However, it binds in similar proportions to the mass of a protein implying the mass of proteins remains unaltered. Since all proteins contain equal negative charges, their separation exclusively relies on the molecular weight of proteins. The result is that proteins with small molecular weight migrate faster than proteins with large molecular proteins (Reginald & Charles, 2010).
The components of the SDS buffer include the chemical buffer used for stabilizing the pH value of the gel. The other component is the counterion solution that balances the charge of the buffer solution. The third component is the SDS detergent responsible for denaturing proteins and nullifying all the charges present in these proteins. For visualization and processing, a running buffer is needed (Burton, 1997). This buffer contains a tracking dye (bromophenol blue) responsible for identifying the movement of the proteins within the gel. In addition, there is the staining dye used for locating the exact position of the bands. Finally, to increase the density of the running samples, additives such as sucrose and glycerol are used to ensure proper movement of the proteins within the gel.
Staining is an important visualization technique used in biochemistry to visualize molecules being separated. An example of a dye used in visualization is Coomassie Blue that binds to molecules such as proteins through nonionic or Van der Waal interactions. The dye contains sulfonic acid groups that bind to amine groups found in proteins. Though it is a less sensitive dye, it is quite convenient because of the ease of its application in protein staining (Schleicher, 2003). This dye appears as dark blue-black at pH 7 but upon acidification turns into a clear tan. However, since the gel is at neutral pH, it retains its blue color and helps in the identification of these bands. The other staining technique that can be used to identify proteins after electrophoresis is silver staining. Using silver nitrate as a major component, it offers excellent sensitivity upon visualization. The rationale behind this staining involves the binding of silver ions to protein molecules to create fine visible images on the gel (Kittelberger, 2000).
Given the complex nature of carrying out silver staining and Coomassie staining, it becomes quite difficult to purify specific proteins. Thus, scientists came up with western blotting as a technique of studying specific proteins after SDS-PAGE electrophoresis. This technique involves the transfer of proteins from the surface of SDS gel to another surface such as polyvinylidene or nitrocellulose. Initially, the transfer of proteins relied upon capillary action but currently the process occurs through the use of electric currents. After the transfer of proteins to the new surface, these proteins can be probed through the use of various reagents (Biji & Hal, 2006). The most commonly used technique is western blotting that involves the use of primary antibodies to bind proteins.
Remember that antibodies bind to proteins in a typical enzyme key and lock system implying that these interactions are specific in nature. After this, secondary antibodies are added to the surface since they bind specifically to certain primary antibodies. These secondary antibodies initiate a luminescent reaction via horseradish peroxidase that emits light to indicate a reaction. These illuminating bands will indicate the presence of proteins separated during gel electrophoresis (Freifelder, 2008). The purpose of these laboratory practicals was to carry out protein separation using SDS-PAGE electrophoresis and then staining the gels using the three staining techniques. In addition, it will also involve calculating the mass of the separated proteins in the gel.
Methodology
The methodology involved in running the gel electrophoresis involves a series of steps. After loading the electrophoretic tank with gel and the combs, let the tank settle for about 20 minutes. After that carefully remove the combs to ensure the created wells do not collapse. Connect the tank to an electric current but do not switch it on. Load the samples carefully into the wells as follows: lane 1-protein marker, lane 2-water, lane 3-PBS, lane 4-PBS plus 1% Triton X-100 and lane 5-PBS plus 2% SDS. After this, switch on the power and let the current run for 30 minutes at the required voltage until the tracking dye runs out of the gel. Switch off the current, pour off the buffer and remove the gel from the tank. The gel is now ready for both silver staining and Coomassie staining.
For Coomassie staining, place the gel in a Petri dish full of distilled water and rinse it for five minutes. Replace the distilled water with the Coomassie stain and let it stain the gel for 30 minutes. Destain the gel with DI water, rinse the gel and then refill it with DI water again. Scan the gel to reveal the bands. After preparing the second gel for silver staining, the actual process of silver staining is as follows: Soak the gel in the staining solution for about 15 minutes. After this, rinse the gel in 100 ml water for 15 minutes twice. After rinsing, soak the same gel in the developing solution for between 2 and 15 minutes. The development is then stopped by rinsing the gel 3times in 7% HAC solution. The gel is then scanned and the bands identified.
Results
From the staining results, it is quite evident that the molecules did not run well on the gel. This may be due to the use of highly concentrated SDS during the denaturation process. As such, these proteins became considerably heavy and during separation, there was a lot of overlapping of the proteins resulting in these unclear images (Matsudaira, 1993). The outcome of the SDS-PAGE is as follows:
The above SDS-PAGE gel was then subject to both silver and Coomassie staining. The scanned results of the gels obtained from the Coomassie staining and silver staining are as follows:
Coomassie staining
<src=”/blog/wp-content/uploads/2012/04/21-225×300.png” alt=”” width=”225″ height=”300″ />
Silver staining
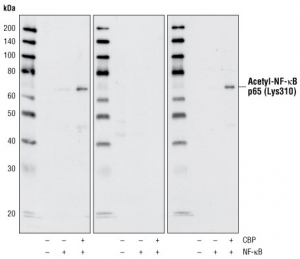
Western blot-likely image
Discussion
Rf refers to the relative mobility of proteins on the gel during electrophoresis. It is the divisor between the migration distance of a band in a single lane and the migration distance of dye front (Szarejko, 2003).
Therefore; Rf = Band Distance/Dye front Distance
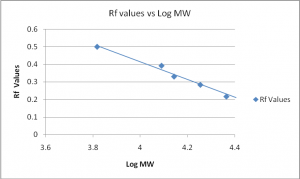
The relative Rf of the five bands as detected by the Coomassie staining is as follows:
Standard weight | Log MW | Rf |
23130 | 4.364 | 0.216 |
17,950 | 4.254 | 0.283 |
13,900 | 4.143 | 0.330 |
12,300 | 4.090 | 0.392 |
6557 | 3.818 | 0.500 |
Table 1
Graph 1
For the silver staining, the standard weights and Rf are as follows:
Standard weight | Log MW | Rf |
17,950 | 4.254 | 0.283 |
13,900 | 4.143 | 0.392 |
12,300 | 4.090 | 0.500 |
2027 | 3.307 | 0.541 |
500 | 2.699 | 0.766 |
Table 2
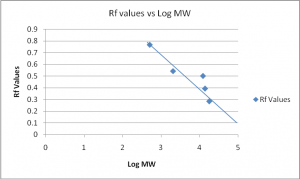
Graph 2
From the western blot technique, three primary protein contents of bovine liver obtained were acting, BSA and GADPH. In terms of Rf and standard weights obtained, the results were as below:
Standard weight | Log MW | Rf |
66,300 | 4.821 | 0.201 |
41,900 | 4.622 | 0.243 |
36,950 | 4.568 | 0.296 |
Table 3
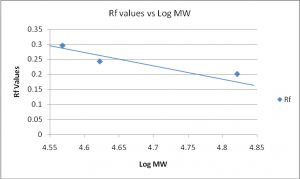
Graph 3
From the results provided after scanning, it is quite clear that the two staining techniques used differ extremely in terms of sensitivity. The ionic interactions between the protein amines and the sulfonic acid groups in Coomassie staining make visualization possible. Similarly, the silver staining reaction also allows visualization of the bands. However, it is quite clear that the silver staining is more sensitive than the Coomassie staining (Moreno, 1999). A comparison of the bands identified using both stains indicates that visibility is more enhanced in silver staining than in Coomassie staining. From the scanned gels, it is evident that both staining techniques do not reveal same proteins. Data in the two tables above show that not all protein bands identified via Coomassie staining will also be reflected in silver staining. As a matter of fact, the two techniques only share three proteins among the identified bands which both have the same Rf values (Burton, 1997). This may be because of the different interaction techniques of the contents of these stains. In addition, Coomassie staining has very low sensitivity when compared with silver staining. This low sensitivity may be responsible for the identification of high molecular weight proteins within the gel effectively. On the other hand, low molecular weight proteins require stains that are highly sensitive to be visualized properly.
Using the standard molecular weights obtained from the listed tables, we can easily identify the proteins in the gel by approximation. Though disparities may exist between the actual molecular weight of the protein and the standard weight determined above, it acceptable because this was an experiment and errors was expected (Reginald & Charles, 2010). From Coomassie staining, the estimated weights of the proteins in the identified bands were as follows 23130, 17,950, 13,900, 12,300 and 6557.
For example, the protein with molecular weight 23,130 is likely to be trypsinogen bovine, 17,950 will be beta-lactoglobulin bovine, 13,900 alpha-lactalbumin bovine, 12,300 is Cytochrome c and 6557 is likely to be aprotinin (Albert, David, & Michael, 2005). The different protein bands identified through silver staining weigh 2027 and 500 Daltons respectively. The two proteins are the S-100 proteins and BLA I proteins found in bovine liver.
During western blotting, three different proteins were identified namely actin, GADPH and BSA proteins. First of all, the three different proteins differ in terms of solubility with GADPH being the most soluble and BSA being the least soluble in both SDS and Triton. The solubility of GADPH may be due to its low molecular weight when compared to the other two that have huge molecular weights. The real weight of GADPH is 37,000 Daltons; actin weighs 42,000 Daltons while BSA weighs 66,000 Daltons (Freifelder, 2008).
In terms of their cellular roles, the three proteins differ substantially in their functions. For instance, actin is a globular protein found in almost all eukaryotic cells. It is a structural protein responsible for the formation of the eukaryotic cytoskeleton. As a result, it offers cells mechanical and physical support, provides trafficking routes during signal transduction and also encourages mobility of cells (Reginald & Charles, 2010). Actin is not a membrane bound protein because it is widespread throughout the cytoplasm. GADPH is a cellular enzyme responsible for the catalyzation of the sixth step in the glycolytic pathway responsible for the generation of ATP. Since the glycolytic process takes place in the cytosol, GADPH is a cytosolic protein. Finally, BSA, the largest of the three proteins identified via western blot, has very limited biological roles in the cells. However, the two major cellular roles where bovine serum albumin makes a contribution are in the mediation of lipid oxidation and lipid binding (Freifelder, 2008).
During the laboratory practical, it is important to recognize that some errors occurred during the preparation of the gel for electrophoresis. The fact that student worked in groups created a lot of disorganization and confusion because of the anxiety of running experiments independently without the presence of the lecturer. However, it should be noted that the absence of the teacher contributed minimally to these errors since the laboratory manual explained every procedure in detail. Personally, I used another gel from a group that did well in terms of staining and SDS preparation. In my case, the source of errors may be attributed to the use of concentrated SDS (more than 2%) due to measurement errors. The numbers of washes after the staining were also not adhered to keenly resulting into poor images.